| ID | hsa_circ_0000816 | |||||
| Alias | hsa_circ_001758 | |||||
| Position | chr17:80521229-80526077 | |||||
| Strand | + | |||||
| Genomic length | 4848 | |||||
| Spliced sequence length | 343 | |||||
| Annotation |
| |||||
| Repeats | None | |||||
| Best transcript | NM_004514 | |||||
| Gene symbol | FOXK2 | |||||
| Scores | circRNA study | Sample | total reads circular junction |
unique reads circular junction |
unique reads linear-5' junctions |
unique reads linear-3' junctions |
|---|---|---|---|---|---|---|
| Maass2017 | adipose | 2 | 2 | 4 | 4 | chondrocytes | 4 | 4 | 2 | 5 | WAS2 | 3 | 3 | 8 | 14 | right_atrium | 2 | 2 | 5 | 5 |
| Jeck2013 | Hs68_RNase | 487 | 158 | 5 | 67 | Hs68_control | 17 | 12 | 30 | 41 |
| Memczak2013 | cd_34 | 5 | 3 | 3 | 1 | HEK293 | 3 | 3 | 2 | 1 | cd_19 | 1 | 1 | 10 | 2 |
| Rybak2015 | SY5Y_exp2_D0 | 7 | 6 | 20 | 26 | cerebellum | 79 | 62 | 53 | 75 | Sy5y_exp1_D0 | 12 | 12 | 38 | 36 | frontal_cortex | 202 | 76 | 76 | 106 | parietal_lobe | 66 | 41 | 65 | 100 | temporal_lobe | 79 | 38 | 28 | 63 | Sy5y_exp1_D2 | 16 | 12 | 27 | 37 | occipital_lobe | 84 | 54 | 54 | 71 | SY5Y_exp2_D4 | 2 | 2 | 18 | 25 | Sy5y_exp1_D4 | 8 | 8 | 17 | 42 | diencephalon | 140 | 97 | 66 | 103 |
| Salzman2013 | Hsmm | 48 | 8 | 9 | 20 | K562 | 127 | 87 | 55 | 107 | Mcf7 | 88 | 21 | 5 | 69 | Nhlf | 45 | 9 | 4 | 17 | Gm12878 | 167 | 77 | 57 | 54 | Sknshra | 66 | 38 | 55 | 62 | H1hesc | 51 | 27 | 65 | 65 | Bj | 41 | 22 | 36 | 29 | Nhek | 98 | 40 | 78 | 82 | Huvec | 20 | 7 | 39 | 67 | Helas3 | 69 | 46 | 117 | 87 | Hepg2 | 98 | 44 | 93 | 83 | A549 | 97 | 47 | 47 | 68 | Ag04450 | 43 | 28 | 30 | 55 |
| Aligned reads | Memczak 2013 circRNA study (pooled samples)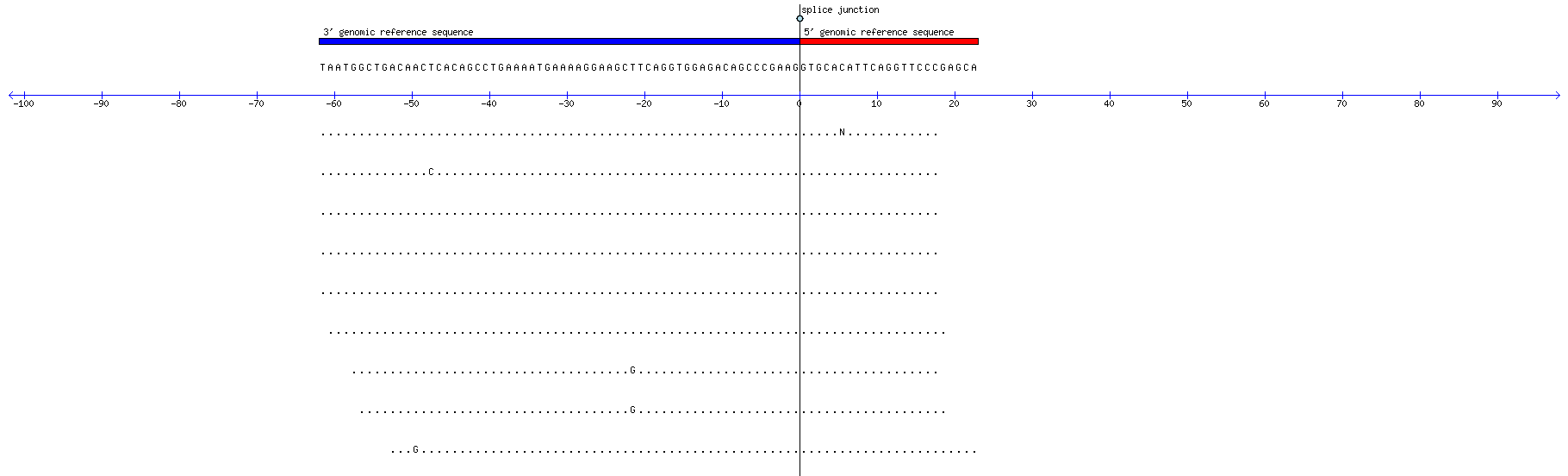 | |||||